Cancer and metabolism-related diseases have a significant impact on humans. Cancer cells are constitutively anabolic. It will cause quiescence and catabolism in normal cells because of they are unable to tolerate nutrient stress. So, oncogenic mutations will reselect the metabolic circuitry during the tumorigenic process to meet the increased anabolic demands of cancer cells. Generally, cancer cells depend on a steady influx of nutrients via cell surface transporters and receptors. Meanwhile, they also can depend on the lysosomal degradation of internalized macromolecules into subunits that can be used for biosynthesis and/or the production of ATP.
Previous reports indicated that endogenous and synthetic sphingolipids can starve many different cancer cell types to death. This is because it can trigger the downregulation of multiple nutrient transporters and/or block lysosome fusion reactions. Furthermore, the sphingolipid analog robustly and acutely blocks ceramide-induced mitochondrial dysfunction, correcting diet-induced obesity and its metabolic sequelae. For example, several sphingolipids can activate protein phosphatase 2A (PP2A) and negatively regulate multiple signaling pathways that promote nutrient transporter expression. Today, we will introduce a sphingolipid analog — SH-BC-893.
SH-BC-893 is an Orally Active Anti-neoplastic Sphingolipid Analog.
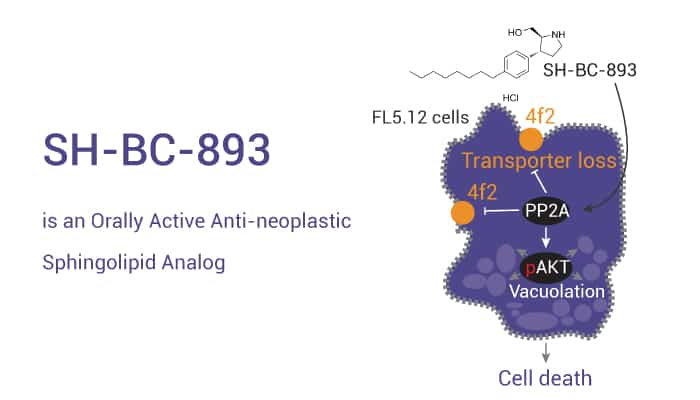
On the one hand, SH-BC-893 is an orally active anti-neoplastic sphingolipid analog. SH-BC-893 starves cancer cells to death by down-regulating cell surface nutrient transporters and blocking lysosomal trafficking events. On the other hand, SH-BC-893 (5 μM; 3 h) protects from ceramide-induced mitochondrial dysfunction by disrupting intracellular trafficking in vitro. And SH-BC-893 (oral; 120 mg/kg; single) also robustly and acutely blocks ceramide-induced mitochondrial dysfunction, correcting diet-induced obesity and its metabolic sequelae in vivo.
In sum, SH-BC-893 is an orally active sphingolipid analog that can be used for research on cancer and obesity.
Reference:
[1] Kubiniok P, Finicle BT, Piffaretti F, et al. Mol Cell Proteomics. 2019;18(3):408-422.
[2] Jayashankar V, Selwan E, Hancock SE, et al. EMBO Mol Med. 2021;13(8):e13086.