Histone acetylation and deacetylation are key events in the regulation of chromatin structure. Additionally, Histone acetyltransferases (HATs) can catalyze acetyl groups to the ε-amino terminus of lysine residues within histones. Acetylation results in an open chromatin structure by removing positive charges from histones. As a result, it induces protein conformational changes. This will lead to transcriptional activity promotion.
Tip60 (KAT5) is a member of the MYST family of HAT enzymes. Loss of Tip60 may lead to impaired DNA repair. The ionizing radiation activates HAT, causing acetylation of histones and activation of p53 and ATM.
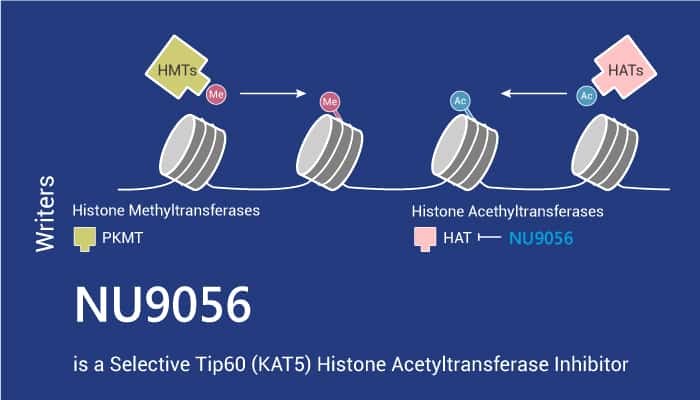
In this article, we will introduce a potent Tip60 (KAT5) histone acetyltransferase inhibitor, NU9056.
NU9056 inhibits Tip60 (KAT5) histone acetyltransferase with an IC50 of 2 nM. What’s more, it shows >16-fold selectivity for Tip60 over PCAF, p300, and GCN5.
In LNCaP cells, NU9056 results in both caspase 3 and caspase 9 activation in a time- and concentration-dependent manner. Besides, NU9056 treatment results in a decreased level of acetylated histone H4K16, H3K14, and H4K8. This all targets Tip60-mediated acetylation. Furthermore, NU9056 treatment also decreases androgen receptor, prostate-specific antigen, p53, and p21 protein levels.
Nextly, in vivo, the mice are injected with Nu9056 (2 μg/g) and the hippocampus is collected 1 h later. Tip60 inhibition reduces H2A.Z binding at the -1 nucleosome of Arc, and the nucleosome of Arc and Syp. Additionally, Nu9056 increases acetylation at the -1 nucleosome of Fos, Tacstd2, and Gria4, and the nucleosome of Gria4.
Protein acetylation is a kind of regulatory mechanism and plays an important role in many signal pathways. The two enzymes HATs and HDACs are de-regulated in disease states. Therefore, targeting both types of enzymes with small molecule inhibitors is a potential way for therapeutic strategy.
Tip60 is a HAT enzyme and plays a particular role in CaP development and progression. And, NU9056 acts as a potent TIP60 inhibitor that has the potential for prostate cancer research.
Reference:
[1]. Kelly Coffey, et al. PLoS One. 2012;7(10):e45539.